Transcriptional circuitry as the achilles heel of Acute myeloid leukemia
Cellular heterogeneity is a major problem in cancer therapy, as treatment-resistant cells can promote relapse. This is a particular problem for malignancies with inherent genetic heterogeneity like Acute Myeloid Leukemia (AML). The advent of single-cell genomics techniques allows the determination of epigenetic and transcriptional profiles of these treatment-resistant populations. This can identify master transcription factors driving the cell fate of different populations within a tumour. Understanding the mechanisms of these master transcription factors and their associated proteins can yield new avenues for therapy to target the entire tumour population, or in combination with existing therapies.
Patient-specific targeting of chromatin regulators in aml
Driver mutations in epigenetic regulators such as DNMT3A, BCOR, EZH2, ASXL1 and TET2 occur in over 40% of Acute Myeloid Leukemias. The effect of these individual mutations on chromatin architecture and transcription is incompletely understood. Using in vitro and clinical models we are uncovering the mechanisms of these chromatin regulators in order to match their mutations to appropriate therapeutics to restore a healthy chromatin state. The rapid expansion of inhibitors and PROTACs for manipulating the epigenome therapeutically provides an exciting avenue for pairing drugs to patient specific mutations.
HISTONE MODIFICATIONS IN TRANSCRIPTIONAL REPRESSION
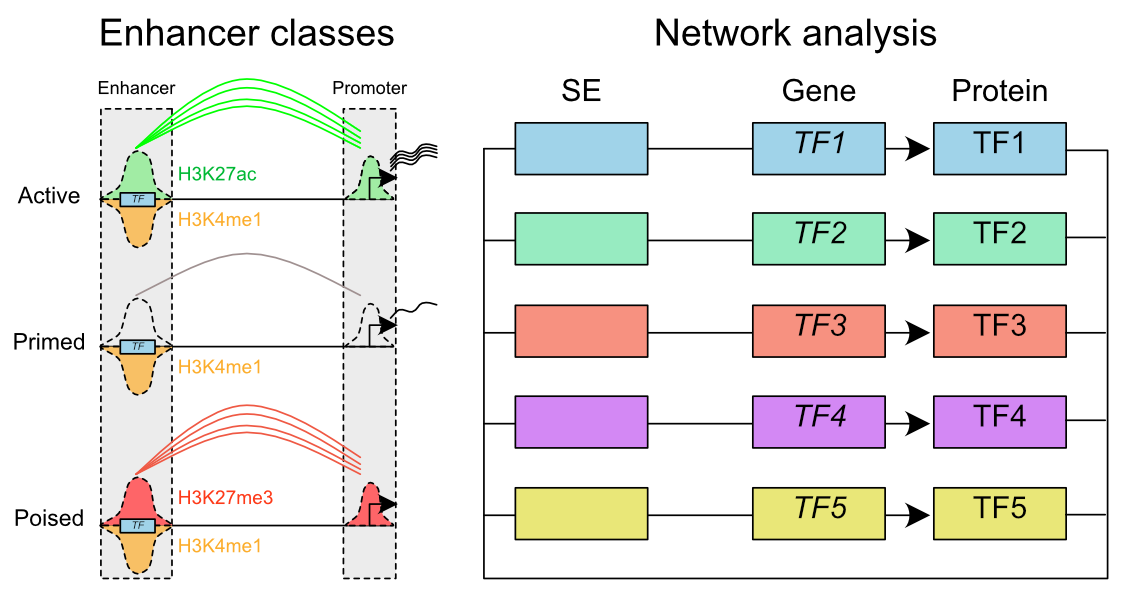
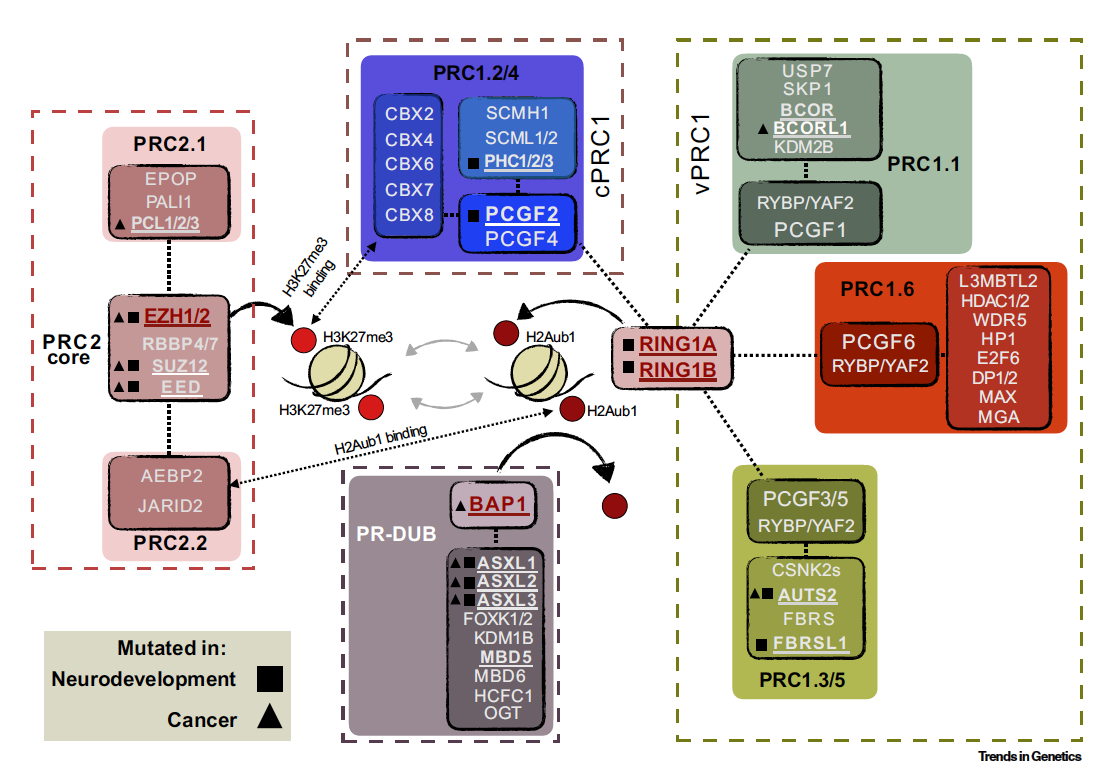
The contribution of histone modifications to transcription has been an area of intensive study over the last decade. It is now evident that the catalytic activity of PRC1 complexes, which mono-ubiquitinate Histone H2A at lysine 119, has a direct role in repression of Polycomb target genes. The mechanism of this repression is not fully understood however, with an apparent disconnect between PRC2 activity and maintenance of repression and a burgeoning list of H2AK119ub1 readers.
Chromatin architecture TECHNIQUES
-
STORM

-
Hi-C and Hi-ChIP

-
ChIP-seq and CUT&Tag

Contact us: eric.conway@ucd.ie
School of Biomolecular and Biomedical Sciences,
University College Dublin, Belfield, Dublin 4